Research Interests
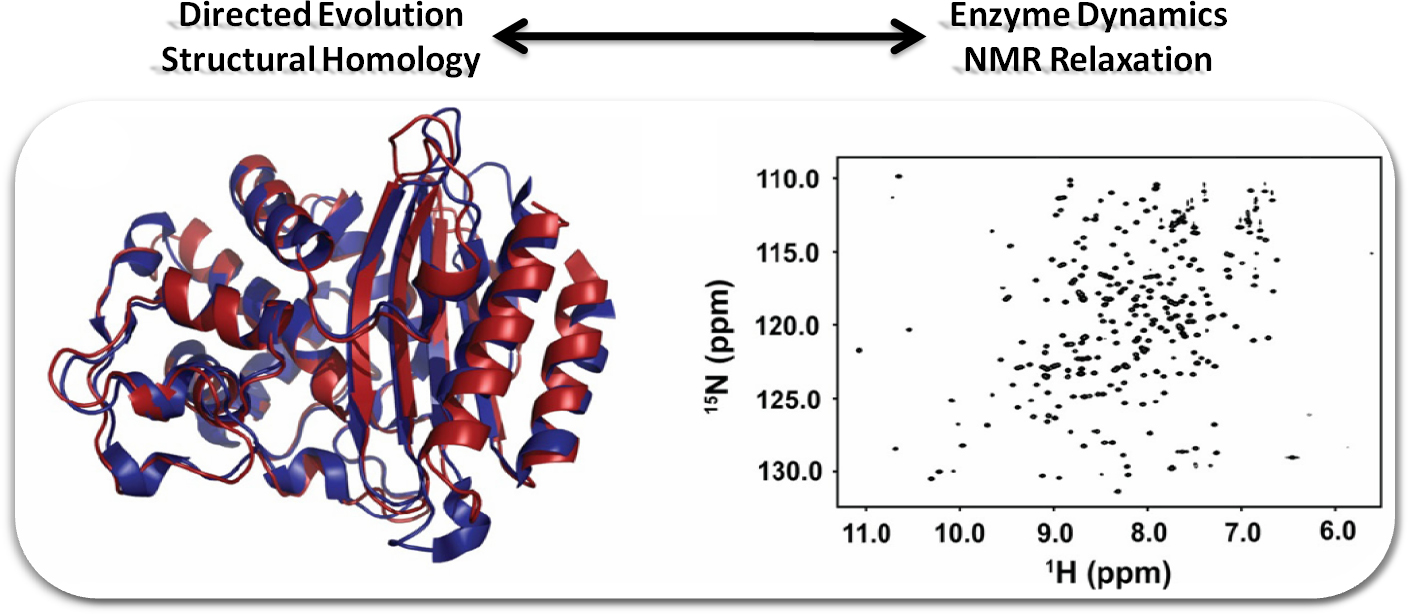
Understanding the importance of residue motion in enzyme activity may eventually allow us to develop methods to modify, modulate and/or design new protein functions, which would have far-reaching implications in biotechnology, protein engineering, nanotechnology and drug design.
The main goal of our research is to successfully modify enzyme biocatalysts aimed at environmental and pharmaceutical applications, with a broader interest focused on understanding the role between structure, function and flexibility in various enzymatic systems.
To address this important problem, our research strategy combines directed evolution and semi-random mutagenesis experiments coupled with biochemical and biophysical characterization such as nuclear magnetic resonance (NMR), molecular biology, enzyme kinetics, and molecular modeling. Using this combined approach, we specifically focus on achieving the following aims:
1) To understand how the amino acid sequence and local structural environment of conserved residues determine the motional and functional properties of enzymes.
2) To investigate how functionally and structurally related enzymes with similar folds retain their dynamical signatures.
3) To modify, reproduce and apply this 'structure-function' and 'flexibility-function' information to the design of new and improved molecular biocatalysts.
Research Funding
We are very grateful to the following sponsors:


 Contact
Info:
Contact
Info: